I am willing to bet most of the people that come across this blog remember a time when you took organic chemistry. While I do not find this topic particularly interesting, mass spectrometry was briefly touched upon. This is an analytical tool that measures the mass of different molecules within a sample by measuring their mass to charge ratio. It allows researchers to know the structure of a molecule with fairly good precision. This tool takes advantage of both the mass and charge of molecules. When ions enter the electric field, they are deflected and this deflection is based on both the mass (lighter ions are deflected more than heavier ions) and charge (more positive ions are deflected more). Why talk about this? Because this tool is actually being employed in microbiology labs throughout the country. You may have heard of it under a different name: MALDI-TOF
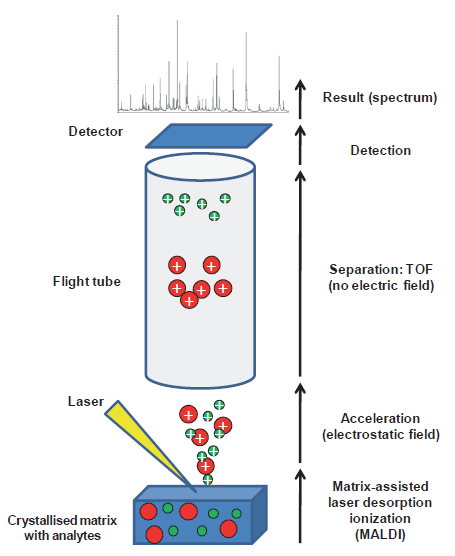
This stands for Matrix Assisted, Laser-Desorption Ionization, Time of Flight. Ok this does not make much sense, but hopefully by explaining the mechanism by which this work, it will make sense.
The technique for MALDI-TOF relies on the generation of positive ions from proteins (1), which are then separated as they moved through a vacuum. There are several ionization techniques, of which 2 are generally used: electron spray ionization (ESI), and matrix assisted-laser desorption ionization (MALDI). Both of these are “soft ionization” methods where ion formation does not lead to loss of sample integrity (2). MALDI is the one that is typically used because it produces singly charged ions (so only +1) so you can measure based on size, and it does not require prior separation by chromatography as for ESI (2). So that takes care of the laser desorption ionization. The matrix assisted part comes from how the sample is prepared for analysis. Samples are prepared by mixing samples with a matrix resulting in crystallization of the sample within the matrix as the sample dries (2,3). The composition of the matrix varies depending to the biomolecule to be analyzed and the type of laser used (3). In general, intact microorganisms can be directly processed, with resistant organisms (spores, viruses, yeast) having to have strong organic acids or alcohols added to the pre-steps. From here, the technique goes as follows:
- Mixture of crystalized matrix is placed on a conductive metal plate
- Brief laser pulses bombards the matrix, which absorbs energy leading to the desorption of the analyzes, which are vaporized and ionized in the gas phase
- These molecules are then accelerated through an electrostatic field and ejected into a flight tube, which is subjected to a vacuum until they reach the detector
- Because there is no electric field in the tube, the time that is particle reaches the detector is measured (aka time of flight). This depends on the mass and charge. In other words, smaller particle reach the detector faster.
- This results in a spectral signature composed of spikes.
In other words, the sample is placed on a matrix, which absorbs lasers, leading to desorption of ions, and ejected into a flight tube where the time of flight is measured. Got it?
After this, the spectral fingerprint is compared to either a database OR matches biomarker masses to a protean database. In other words, the fingerprint that is obtained from the sample can be compared to previously collected fingerprint libraries, which is the simplest way of getting a result. If an unknown organism is identified, then the protein molecular masses on the spectrum are matched with protein molecular masses predicted from sequenced genomes (3). An algorithm then predicts protein masses from the genome and seeks matches with experimentally derived masses. Caveat here is that it can only be applied to organisms whose genome has been sequenced. What does this look like and what do we, as clinicians, get back? This is a fingerprint for both E.coli and Shigella (4):
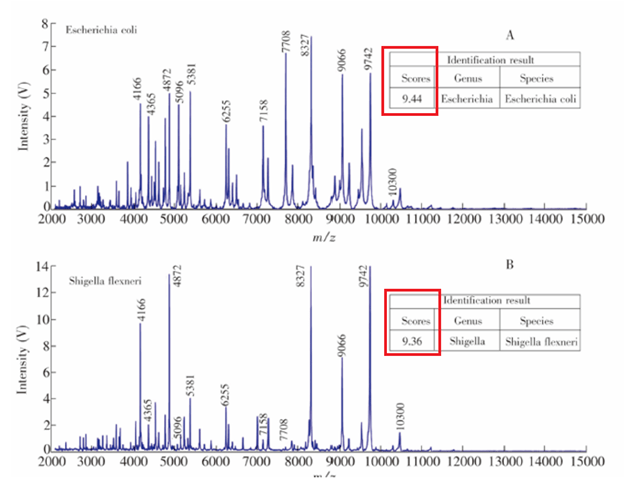
You may have noticed that scoring system up above. What is this all about? This varies depending on the manufacturer (5). In other words, the specific hardware use will look at the fingerprint, run an algorithm, and spit out a score that will let you know the species consistency that is based on their database and their algorithm. High values represent high similarity with respective database entries. The quality of species identification is divided into several categories, depending on the score. For instance, the Brunker score assign a score >1.9 for identification to species and genus level, while score >1.7 being that of the same genus. Those who are <1.7 are regarded as unreliable. Again, these scores will vary based on the manufacturer and the algorithm is not released to the public.
Data Favoring MALDI-TOF
Many studies have compared the accuracy of MALDI-TOF to that of standard microbiological methods. For instance, one study evaluated 697 isolates of staphylococci (5) and found that 97.3% of isolates were identified correctly to the species level by MALDI-TOF. The exception to this was S. xylosus. In another study of 133 positive blood cultures (6), MALDI-TOF found that 95.9% of specimens had a score >2 (definite identification of genus and probable identification to species level) for gram negative bacteria. The same study found concordance for 90% of gram positive cocci. This pattern held true even in a study of 611 isolates across different identification methods and types of organisms (7):
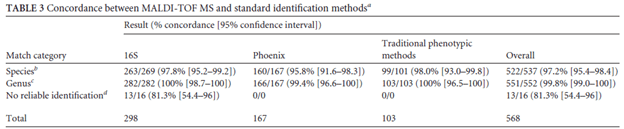
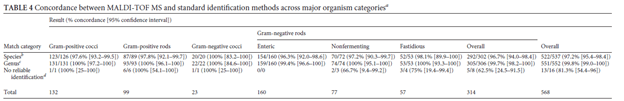
MALDI-TOF has also been used to identify mycobacteria and fungi. For instance, a study of 167 species found that MALDI-TOF identified 87% of the rapid growing NTM, 75% of slow growing NTM, and 100% of MTB. Indeed, another study of 61 positive cultures found that the turn around time for conventional identification was 40.9h compared to 6.6h for MALDI-TOF.
While out of the scope, MALDI-TOF has been used to identify fungi and viruses (2,3) and this modality will become more prevalent as we go forward, given its low turnaround time for identification. Despite these advantages, we need to remember that MALDI-TOF depends on the quality of the databases as well as hardware. Despite this, MALDI-TOF remains a valuable tool for microbiological diagnosis going forward.
References:
- Bennett, J. E., Dolin, R., & Blaser, M. J. (2014). Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Elsevier Inc.
- Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. 2015 Aug 5;6:791. doi: 10.3389/fmicb.2015.00791. PMID: 26300860; PMCID: PMC4525378.
- Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012 Mar;36(2):380-407. doi: 10.1111/j.1574-6976.2011.00298.x. Epub 2011 Aug 22. PMID: 22092265.
- Yu, Jiajun & LIU, Ping & ZENG, Zhen & CHEN, Ying & Gao, Wei & LI, Mei & WANG, Chen-Guang & HUANG, Zheng-Xu & Zhou, Zhen & Lei, Li. (2018). Development and Characterization of A Linear Matrix-assisted Laser Desorption Ionization Mass Spectrometer. Chinese Journal of Analytical Chemistry. 46. 463-470. 10.1016/S1872-2040(17)61077-6.
- Richter C, Hollstein S, Woloszyn J, Kaase M, Gatermann SG, Szabados F. Evaluation of species-specific score cut-off values for various Staphylococcus species using a MALDI Biotyper-based identification. J Med Microbiol. 2012 Oct;61(Pt 10):1409-1416. doi: 10.1099/jmm.0.042606-0. Epub 2012 Jun 28. PMID: 22745134.
- Barnini S, Ghelardi E, Brucculeri V, Morici P, Lupetti A. Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol. 2015 Jun 18;15:124. doi: 10.1186/s12866-015-0459-8. PMID: 26084329; PMCID: PMC4471905.
- Khot PD, Couturier MR, Wilson A, Croft A, Fisher MA. Optimization of matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis for bacterial identification. J Clin Microbiol. 2012 Dec;50(12):3845-52. doi: 10.1128/JCM.00626-12. Epub 2012 Sep 19. PMID: 22993178; PMCID: PMC3502975.
- Alcolea-Medina A, Fernandez MTC, Montiel N, García MPL, Sevilla CD, North N, Lirola MJM, Wilks M. An improved simple method for the identification of Mycobacteria by MALDI-TOF MS (Matrix-Assisted Laser Desorption- Ionization mass spectrometry). Sci Rep. 2019 Dec 27;9(1):20216. doi: 10.1038/s41598-019-56604-7. PMID: 31882826; PMCID: PMC6934676.
- Lagacé-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, Webb AA, Miller C, Alfa MJ. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012 Oct;50(10):3324-8. doi: 10.1128/JCM.01479-12. Epub 2012 Aug 8. PMID: 22875888; PMCID: PMC3457416.

1 comments on “MALDI-TOF – TARDIS?”